Reading Parquet Files
Syntax
help(spark.read.parquet)
df = spark.read \
.format("parquet") \
.option("mergeSchema", "true") \ # Merges schemas of all files (useful when reading from multiple files with different schemas)
.option("pathGlobFilter", "*.parquet") \ # Read only specific files based on file name patterns
.option("recursiveFileLookup", "true") \ # Recursively read files from directories and subdirectories
.load("/path/to/parquet/file/or/directory")
Options
- mergeSchema: When reading Parquet files with different schemas, merge them into a single schema.
- true (default: false)
- pathGlobFilter: Allows specifying a file pattern to filter which files to read (e.g., “*.parquet”).
- recursiveFileLookup: Reads files recursively from subdirectories.
- true (default: false)
- modifiedBefore/modifiedAfter: Filter files by modification time. For example:
.option(“modifiedBefore”, “2023-10-01T12:00:00”)
.option(“modifiedAfter”, “2023-01-01T12:00:00”) - maxFilesPerTrigger: Limits the number of files processed in a single trigger, useful for streaming jobs.
- schema: Provides the schema of the Parquet file (useful when reading files without inferring schema).
from pyspark.sql.types import StructType, StructField, IntegerType, StringTypeschema = StructType([StructField("id", IntegerType(), True), StructField("name", StringType(), True)])
df = spark.read.schema(schema).parquet("/path/to/parquet")
Path
- Load All Files in a Directory
df = spark.read.parquet(“/path/to/directory/”) - Load Multiple Files Using Comma-Separated Paths
df = spark.read.parquet(“/path/to/file1.parquet”, “/path/to/file2.parquet”, “/path/to/file3.parquet”) - Using Wildcards (Glob Patterns)
df = spark.read.parquet(“/path/to/directory/*.parquet”) - Using Recursive Lookup for Nested Directories
df = spark.read.option(“recursiveFileLookup”, “true”).parquet(“/path/to/top/directory”) - Load Multiple Parquet Files Based on Conditions
df = spark.read .option(“modifiedAfter”, “2023-01-01T00:00:00”) .parquet(“/path/to/directory/”) - Programmatically Load Multiple Files
file_paths = [“/path/to/file1.parquet”, “/path/to/file2.parquet”, “/path/to/file3.parquet”]
df = spark.read.parquet(*file_paths) - Load Files from External Storage (e.g., S3, ADLS, etc.)
df = spark.read.parquet(“s3a://bucket-name/path/to/files/”)
Example
# Reading Parquet files with options
df = spark.read \
.format("parquet") \
.option("mergeSchema", "true") \
.option("recursiveFileLookup", "true") \
.load("/path/to/parquet/files")
Conclusion
To load multiple Parquet files at once, you can:
- Load an entire directory.
- Use wildcard patterns to match multiple files.
- Recursively load from subdirectories.
- Programmatically pass a list of file paths. These options help streamline your data ingestion process when dealing with multiple Parquet files in Databricks.
Write to parquet
Syntax
# Writing a Parquet file
df.write \
.format("parquet") \
.mode("overwrite") \ # Options: "overwrite", "append", "ignore", "error"
.option("compression", "snappy") \ # Compression options: none, snappy, gzip, lzo, brotli, etc.
.option("maxRecordsPerFile", 100000) \ # Max number of records per file
.option("path", "/path/to/output/directory") \
.partitionBy("year", "month") \ # Partition the output by specific columns
.save()
Options
compression: .option(“compression”, “snappy”)
Specifies the compression codec to use when writing files.
Options: none, snappy (default), gzip, lzo, brotli, lz4, zstd, etc.
maxRecordsPerFile: .option(“maxRecordsPerFile”, 100000)
Controls the number of records per file when writing.
Default: None (no limit).
saveAsTable: saveAsTable(“parquet_table”)
Saves the DataFrame as a table in the catalog.
Save: save()
path:
Defines the output directory or file path.
mode: mode(“overwrite”)
Specifies the behavior if the output path already exists.
- overwrite: Overwrites existing data.
- append: Appends to existing data.
- ignore: Ignores the write operation if data already exists.
- error or errorifexists: Throws an error if data already exists (default).
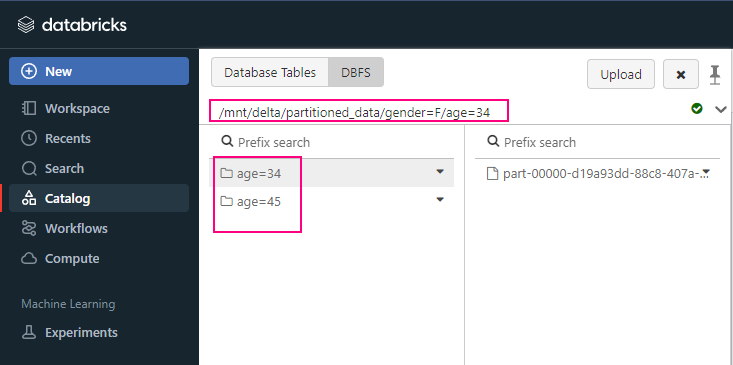
Partition: partitionBy(“year”, “month”)
Partitions the output by specified columns
bucketBy: .bucketBy(10, “id”)
Distributes the data into a fixed number of buckets
df.write \
.bucketBy(10, "id") \
.sortBy("name") \
.saveAsTable("parquet_table")
Example
# Writing Parquet files with options
df.write \
.format("parquet") \
.mode("overwrite") \
.option("compression", "gzip") \
.option("maxRecordsPerFile", 50000) \
.partitionBy("year", "month") \
.save("/path/to/output/directory")
writing key considerations:
- Use mergeSchema if the Parquet files have different schemas, but it may increase overhead.
- Compression can significantly reduce file size, but it can add some processing time during read and write operations.
- Partitioning by columns is useful for organizing large datasets and improving query performance.