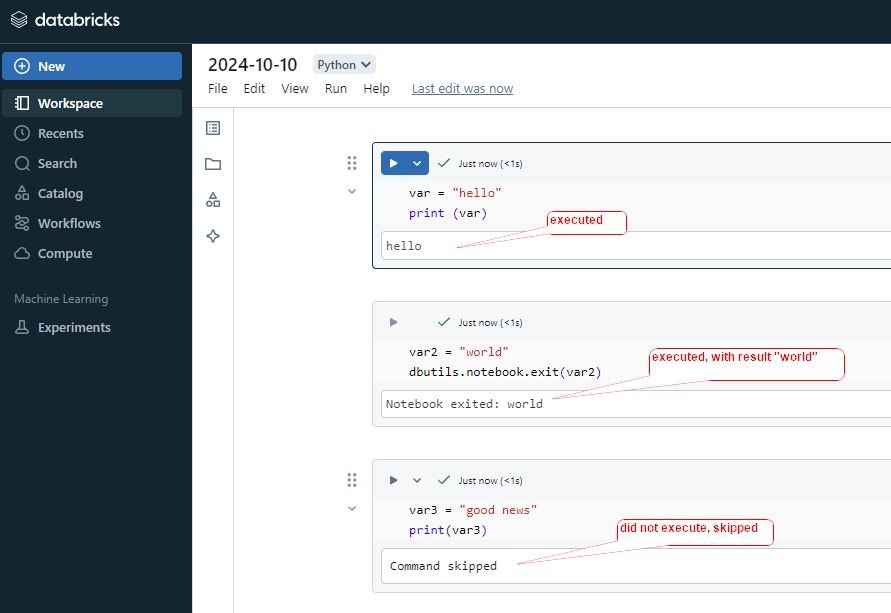
alias ()
alias () is used to assign a temporary name or “alias” to a DataFrame, column, or table, which can be used for reference in further operations
# for dataframe:
df1 = df.alias("df1")
df1.show()
==output==
+---+---+
| id|age|
+---+---+
| 1| 25|
| 2| 12|
| 3| 40|
+---+---+caution: df.alias(“newName”) will not generate new dataframe,
# for column:
df.select(df.id.alias("new_ID")).show()
df.select(df["id"].alias("new_ID")).show()
df.select(col("id").alias("new_ID")).show()
==output==
+------+
|new_ID|
+------+
| 1|
| 2|
| 3|
+------+asc(), desc ()
asc (): ascending order when sorting the rows of a DataFrame by one or more columns.
sample df
+---+---+
| id|age|
+---+---+
| 1| 25|
| 2| 12|
| 3| 40|
+---+---+from pyspark.sql.functions import asc
df.orderBy(asc("age")).show()
==output==
+---+---+
| id|age|
+---+---+
| 2| 12|
| 1| 25|
| 3| 40|
+---+---+desc (): descending order when sorting the rows of a DataFrame by one or more columns.
from pyspark.sql.functions import desc
df.orderBy(desc("age")).show()
==output==
+---+---+
| id|age|
+---+---+
| 3| 40|
| 1| 25|
| 2| 12|
+---+---+cast ()
df[“column_name”].cast(“new_data_type”)
This can be a string representing the data type (e.g., "int", "double", "string", etc.) or a PySpark DataType object (like IntegerType(), StringType(), FloatType(), etc.).
Common Data Types:
IntegerType(),"int": For integer values.DoubleType(),"double": For double (floating-point) values.FloatType(),"float": For floating-point numbers.StringType(),"string": For text or string values.DateType(),"date": For date values.TimestampType(),"timestamp": For timestamps.BooleanType(),"boolean": For boolean values (true/false).
sample dataframe
+---+---+
| id|age|
+---+---+
| 1| 25|
| 2| 12|
| 3| 40|
+---+---+
df.printSchema()
root
|-- id: long (nullable = true)
|-- age: long (nullable = true)from pyspark.sql.functions import col
# Cast a string column to integer
df1 = df.withColumn("age_int", col("age").cast("int"))
df1.printSchema()
==output==
root
|-- id: long (nullable = true)
|-- age: long (nullable = true)
|-- age_int: integer (nullable = true)
# Cast 'id' from long to string and 'age' from long to double
df_casted = df.withColumn("id", col("id").cast("int")) \
.withColumn("age", col("age").cast("double"))
df_casted.show()
df_casted.printSchema()
==output==
+---+----+
| id| age|
+---+----+
| 1|25.0|
| 2|12.0|
| 3|40.0|
+---+----+
root
|-- id: string (nullable = true)
|-- age: double (nullable = true)filter (), where (),
filter () or where () function is used to filter rows from a DataFrame based on a condition or set of conditions. It works similarly to SQL’s WHERE clause,
df.filter(condition)
df.where(condition)
Condition (for ‘filter’)
&(AND)|(OR)~(NOT)- == (EQUAL)
all “filter” can change to “where”, vice versa.
sample dataframe
+------+---+-------+
| Name|Age|Salary|
+------+---+-------+
| Alice| 30| 50000|
| Bob| 25| 30000|
|Alicia| 40| 80000|
| Ann| 32| 35000|
+------+---+-------+
# Filter rows where age is greater than 30 AND salary is greater than 50000
df.filter((df["age"] > 30) & (df["salary"] > 50000))
df.where((df["age"] > 30) & (df["salary"] > 50000))
+------+---+------+
| Name|Age|Salary|
+------+---+------+
|Alicia| 40| 80000|
+------+---+------+
# Filter rows where age is less than 25 OR salary is less than 40000
df.filter((df["age"] < 25) | (df["salary"] < 40000))
df.where((df["age"] < 25) | (df["salary"] < 40000))
+----+---+------+
|Name|Age|Salary|
+----+---+------+
| Bob| 25| 30000|
| Ann| 32| 35000|
+----+---+------+
like ()
like() function is used to perform pattern matching on string columns, similar to the SQL LIKE operator
df.filter(df[“column_name”].like(“pattern”))
Pattern
%: Represents any sequence of characters._: Represents a single character.
pattern is case sensitive.
sample dataframe
+------+---+
| Name|Age|
+------+---+
| Alice| 30|
| Bob| 25|
|Alicia| 28|
| Ann| 32|
+------+---+
# Filtering names that start with 'Al'
df.filter(df["Name"].like("Al%")).show()
+------+---+
| Name|Age|
+------+---+
| Alice| 30|
|Alicia| 28|
+------+---+
# Filtering names that end with 'n'
df.filter(df["Name"].like("%n")).show()
+----+---+
|Name|Age|
+----+---+
| Ann| 32|
+----+---+
# Filtering names that contain 'li'
df.filter(df["Name"].like("%li%")).show()
+------+---+
| Name|Age|
+------+---+
| Alice| 30|
|Alicia| 28|
+------+---+
# Filtering names where the second letter is 'l'
df.filter(df["Name"].like("A_l%")).show()
+----+---+
|Name|Age|
+----+---+
+----+---+
nothing found in this pattern